.braidz files
A .braidz file contains the results of realtime tracking, the tracking
parameters, and so on.
Viewer
A viewer for .braidz files is at braidz.strawlab.org.
Analysis scripts
Scripts to analyze your .braidz files can be found at github.com/strawlab/strand-braid/tree/main/strand-braid-user/analysis.
Latency Analysis
To analyze the latency of your setup with Braid, you can use the Jupyter Notebook braid-latency-analysis.ipynb.
Content analysis
The Jupyter Notebook braidz-contents.ipynb can be used to view the Kalman estimates, the raw 2D detections, data on the association of cameras, and data on associations between 2D detection and 3D Tracking in your .braidz file.
Plotting
The following plots were made with the file
20201112_133722.braidz.
The scripts can be accessed at
github.com/strawlab/strand-braid/tree/main/strand-braid-user/analysis.
A Jupyter Notebook to create all of these plots can be found in braid-plotting.ipynb in the same folder.
braid-analysis-plot-data2d-timeseries.py
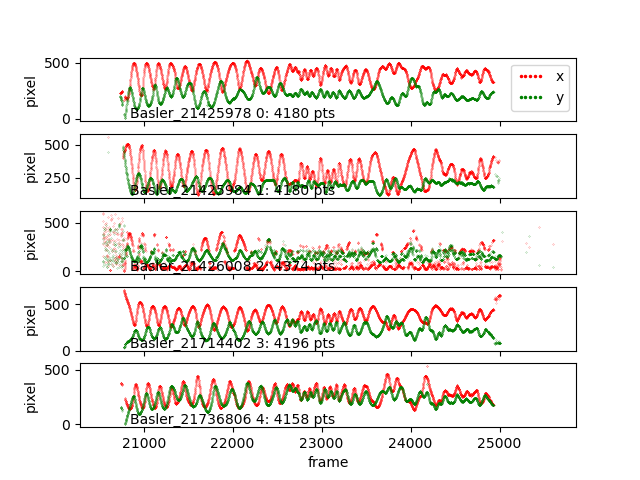
braid-analysis-plot-kalman-estimates-timeseries.py
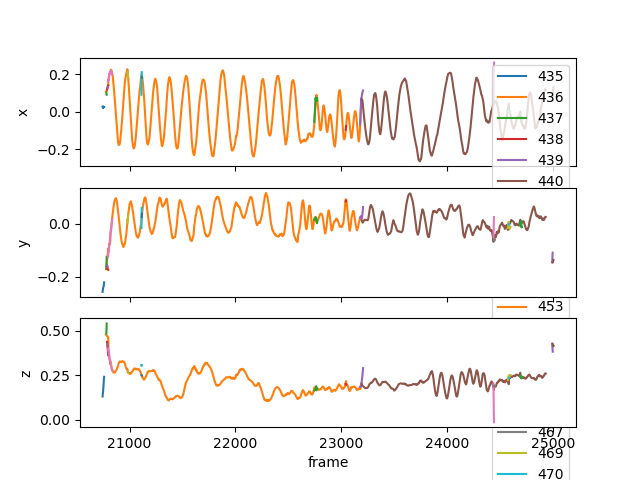
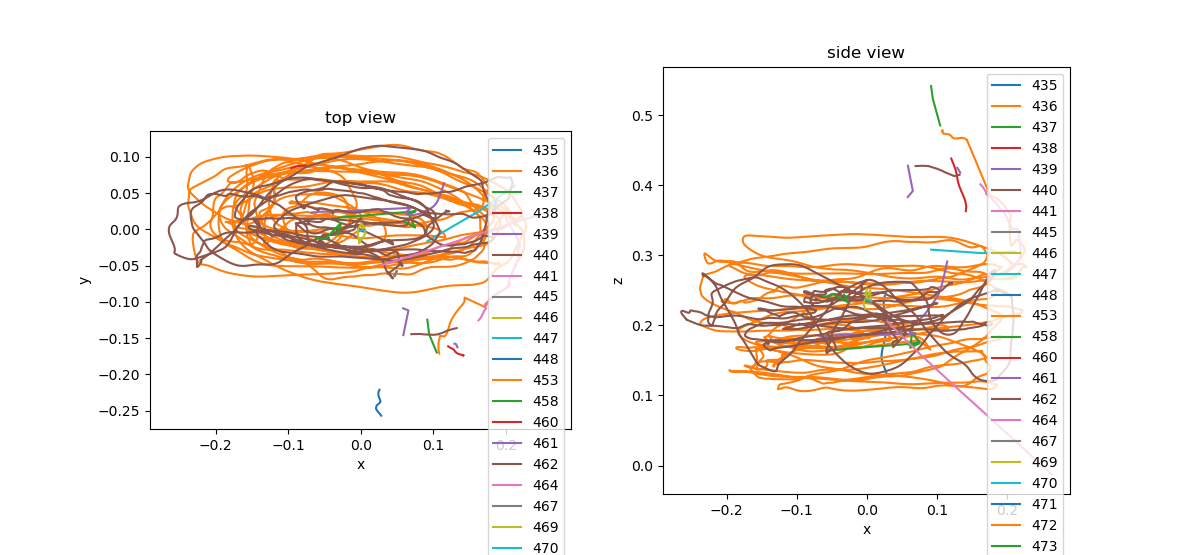
braid-analysis-plot3d.py
File Format
A .braidz file is actually a ZIP file with specific contents. It can be
helpful to know about these specifics when problems arise.
Showing the contents of a .braidz file
You can show the filenames inside a .braidz file with
unzip -l FILENAME.braidz.
Extracting a .braidz file
You can extract a .braidz file to its contents with unzip FILENAME.braidz.
Creating a .braidz file
You can create a new .braidz file with:
cd BRAID_DIR
zip ../FILENAME.braidz *
Note, your .braidz file should look like this - with no directories other than
images/.
$ unzip -l 20191125_093257.braidz
Archive: 20191125_093257.braidz
zip-rs
Length Date Time Name
--------- ---------- ----- ----
97 2019-11-25 09:33 README.md
155 2019-11-25 09:33 braid_metadata.yml
0 2019-11-25 09:33 images/
308114 2019-11-25 09:33 images/Basler_22005677.png
233516 2019-11-25 09:33 images/Basler_22139107.png
283260 2019-11-25 09:33 images/Basler_22139109.png
338040 2019-11-25 09:33 images/Basler_22139110.png
78 2019-11-25 09:33 cam_info.csv.gz
2469 2019-11-25 09:33 calibration.xml
397 2019-11-25 09:33 textlog.csv.gz
108136 2019-11-25 09:33 kalman_estimates.csv.gz
192 2019-11-25 09:33 trigger_clock_info.csv.gz
30 2019-11-25 09:33 experiment_info.csv.gz
2966 2019-11-25 09:33 data_association.csv.gz
138783 2019-11-25 09:33 data2d_distorted.csv.gz
--------- -------
1416233 15 files
Note that the following is NOT a valid .braidz file because it has a leading
directory name for each entry.
$ unzip -l 20191119_114103.NOT-A-VALID-BRAIDZ
Archive: 20191119_114103.NOT-A-VALID-BRAIDZ
Length Date Time Name
--------- ---------- ----- ----
0 2019-11-19 11:41 20191119_114103.braid/
97 2019-11-19 11:41 20191119_114103.braid/README.md
155 2019-11-19 11:41 20191119_114103.braid/braid_metadata.yml
0 2019-11-19 11:41 20191119_114103.braid/images/
320906 2019-11-19 11:41 20191119_114103.braid/images/Basler_22005677.png
268847 2019-11-19 11:41 20191119_114103.braid/images/Basler_22139107.png
308281 2019-11-19 11:41 20191119_114103.braid/images/Basler_22139109.png
346232 2019-11-19 11:41 20191119_114103.braid/images/Basler_22139110.png
225153 2019-11-19 11:41 20191119_114103.braid/images/Basler_40019416.png
86 2019-11-19 11:41 20191119_114103.braid/cam_info.csv.gz
2469 2019-11-19 11:41 20191119_114103.braid/calibration.xml
10 2019-11-19 11:41 20191119_114103.braid/textlog.csv.gz
10 2019-11-19 11:41 20191119_114103.braid/kalman_estimates.csv.gz
10 2019-11-19 11:41 20191119_114103.braid/trigger_clock_info.csv.gz
10 2019-11-19 11:41 20191119_114103.braid/experiment_info.csv.gz
10 2019-11-19 11:41 20191119_114103.braid/data_association.csv.gz
20961850 2019-11-19 12:17 20191119_114103.braid/data2d_distorted.csv.gz
--------- -------
22434126 17 files
Contents of a .braidz file
The most important tables in the .braidz file are kalman_estimates, with the
3D tracking results, and data2d_distorted, with the 2D camera detections.
data2d_distorted table
The data2d_distorted table contains the raw (2D) camera detections and is
typically quite large. See the documentation for the row type
Data2dDistortedRow.
This file is important for carrying synchronization data between cameras. For
example, when saving videos, the timing data carried by the
frame
and
block_id
fields is important.
kalman_estimates table
The kalman_estimates tables contains the estimated state (positions and
velocities) of each tracked object in addition to the estimated covariance. See
the documentation for the row type
KalmanEstimatesRow.
data_association table
The data_association table contains which camera detections contributed to
estimating the state of which objects in the kalman_estimates table. See the
documentation for the row type
DataAssocRow.
Chunked iteration of kalman_estimates
The primary tracking results are in the kalman_estimates table. There can
often be many gigabytes of data here, and thus it is useful to iterate over
duration-defined chunks in this file. This way, the entire .braidz file never
needs to be decompressed and the all results do not need to fit in your
computer's memory at once.
This following example uses the pybraidz_chunked_iter Python package. It
iterates over chunks of the file 20201104_174158.braidz, which can be
downloaded here:
import pybraidz_chunked_iter # install with "pip install pybraidz_chunked_iter"
import pandas as pd
# The filename of the braidz file
braidz_fname = "20201104_174158.braidz"
# Open the braidz file and create chunks of 60 second durations.
estimates_chunker = pybraidz_chunked_iter.chunk_on_duration(braidz_fname, 60)
# One could also create chunks with 100 frames of data.
# estimates_chunker = pybraidz_chunked_iter.chunk_on_num_frames(braidz_fname, 100)
# Iterate over each chunk
for chunk in estimates_chunker:
print("Read chunk with %d rows"%(chunk["n_rows"],))
# Create a pandas DataFrame with the data from each chunk
df = pd.DataFrame(data=chunk["data"])
print(df)